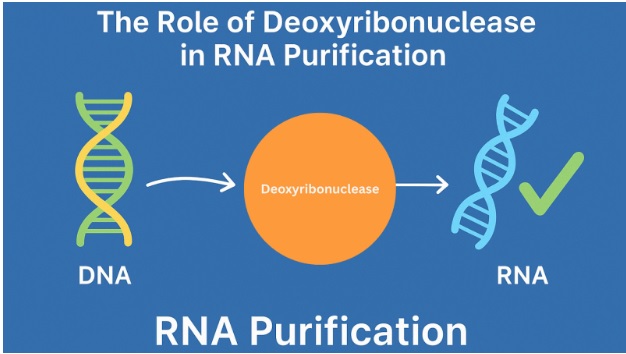
The Role of Deoxyribonuclease in RNA Purification
Modern molecular biology, transcriptomics, and clinical research require high-quality RNA. Even minor impurities can lead to flawed outcomes. Residual genomic DNA is the most common and problematic contaminant present in RNA.
Researchers incorporate Deoxyribonuclease (DNase) treatment in the RNA purification process to address these challenges. Highly purified forms of human deoxyribonuclease can degrade contaminating DNA while maintaining RNA integrity.
Deoxyribonuclease
These are nucleases that cleave phosphodiester bonds to hydrolyze DNA. Thereby, long DNA polymers are converted into shorter fragments or single nucleotides. The following are the two major categories of DNases:
- DNase I
- DNase II
| Feature | DNase I | DNase II |
| Cofactor Requirement | Requires divalent cations | Not required |
| Optimal Conditions | Neutral pH | Acidic pH |
| Preferred Substrate | Double-stranded DNA | Double-stranded DNA |
| Cleavage Pattern | Produces 3′-OH and 5′-phosphate termini | Produces 3′-phosphate ends |
| Use in Research | RNA purification | Lysosomal DNA degradation |
Human deoxyribonuclease, originating from human sources, offers:
- High catalytic efficiency
- Reduced immunogenicity
- RNase-free purity
These deoxyribonucleases, ensuring consistent, specific DNA removal, are valuable for sensitive RNA purification.
Role of DNase Treatment in RNA Purification
Residual DNA in RNA samples can lead to:
- False-positive amplification
- Distorted gene expression measurements
- Reduced sequencing library quality
- Unreliable downstream analyses
- Ultimately leading to compromised conclusions
How DNase improves RNA integrity
Trace amounts of genomic DNA often co-purify with RNA. DNase eliminates the genomic DNA to preserve the true biological integrity of RNA. This helps prevent DNA-derived signals and ensures that true RNA levels are measured.
There is no false-positive amplification in qPCR/RT-PCR/dPCR. Maintaining RNA-seq quality prevents mapping errors, library prep issues, and skewed transcript counts. It also helps avoid background signals that mask rare RNA species.
Methods of DNase Treatment in RNA Purification
The choice of DNase treatment method depends on the following key factors:
- The workflow
- Sample type
- Desired RNA purity
On-column digestion and in-solution treatment are the most commonly used methods.
DNase activity must be terminated properly to prevent RNA degradation. This is achieved via heat inactivation or cleanup methods.
On-column DNase
In this method, DNase is applied directly to RNA bound on the purification column. This method is not only easy to perform but also integrates with standard kits. However, it may not remove heavy DNA contamination.
In-solution DNase
In this method, DNase is added to a solution containing RNA. This is done during or immediately after extraction, before final purification. It is flexible for different amounts, and the enzyme concentration can be optimized accordingly. However, this method requires additional cleanup to remove the enzyme and DNA fragments.
Heat inactivation
This method deactivates DNase by applying a brief heat treatment after digestion. It is fast and straightforward, but has the following disadvantages:
- It may not fully deactivate stable DNase.
- High heat can affect RNA.
DNase Cleanup
This method uses columns, magnetic beads, or precipitation to remove degraded DNA and DNase. While an extra step adds time, it ensures complete removal and preserves RNA quality.
Optimizing DNase Treatment
Optimization is done by carefully balancing enzyme activity, reaction conditions, and RNA stability.
Enzyme Concentration and Incubation Time
Using too little DNase often leaves residual DNA. On the other hand, excessive DNase use can degrade RNA. Researchers should use the manufacturer’s recommended concentrations. The concentration should be adjusted based on RNA amount, DNA contamination level, and sample type. Shorter incubation with sufficient enzymes can minimize RNA exposure to nucleases.
Buffer Composition and Required Cofactors
DNase activity greatly depends on optimal buffer conditions and cofactors. Improper buffers can reduce efficiency or increase nonspecific degradation.
Always use buffers recommended for the specific DNase. Also, ensure:
- Correct pH
- Ionic strength
- Cofactor concentration
Avoid buffers that contain RNases or chelating agents (like EDTA) that inhibit DNase activity.
Temperature Control
While high temperatures accelerate DNase activity and may destabilize RNA, low temperatures slow the reaction. Perform digestion at the recommended temperature and avoid prolonged exposure at elevated temperatures.
Avoiding RNA Degradation
RNA is highly susceptible to degradation by RNases present in reagents or on surfaces. Even optimized DNase treatment can fail if RNA integrity is compromised.
Always use RNase-free reagents, consumables, and gloves. RNase inhibitors should be included where appropriate. Keep handling time to a minimum and terminate DNase activity promptly.
Applications in Research
Gene Expression Studies (qPCR/RT-PCR)
Residual genomic DNA can lead to false-positive amplification in quantitative PCR (qPCR) and reverse transcription PCR (RT-PCR), skewing gene expression results. DNase treatment ensures that the detected signal originates solely from RNA transcripts.
RNA Sequencing (bulk and single-cell)
DNA contamination can lead to inaccurate mapping and distorted transcript counts. It can also compromise library preparation. DNase eliminates contaminating DNA, and the final data reflect the real transcriptome.
Clinical Diagnostics & Biomarker Research
Reliable RNA quantification underpins many clinical applications, including biomarker identification and RNA-based diagnostic panels. DNase treatment removes residual genomic DNA. The data reflects genuine outcomes instead of contamination-driven artifacts.